Table Of Contents
Background Link to heading
To explore Python performance I decided to use the Mandelbrot set. The Mandelbrot set is defined in the complex plane as simply the set of complex numbers that do not diverge as you compute the series $$f(z) = z^2 + c$$. Where c is a complex number. Some numbers will diverge very quickly, where as others could take infinite amount of time, so in practice the number of iterations until we declare a c as being convergent is set. In all of the implementations below I went with 1,000 as the limit. You can set the number of iterations as high as you like, but it will just take longer (to infinitely long). The end goal of this exploration is to create a Mandelbrot set visualizer that automatically zooms, so I didn’t want to set the iterations too high.
The rest of this post will walk through different implementations of computing the Mandelbrot set with a 1200x1200 grid of the complex plane. We will use 1,000 iterations to decide that the set has not diverged. Each run will save off an image of the computed Mandelbrot set.
Simple Python Approach Link to heading
Since the definition of the Mandelbrot set is so simple I started with a naive brute force implementation in Python.
def mandelbrot_naive(xs, ys, _, max_iter, power=2):
output = []
for i in range(len(xs)):
for j in range(len(ys)):
c = complex(xs[i], ys[j])
n = 0
z = 0
while abs(z) < 2 and n < max_iter:
z = z**power + c
n += 1
output.append(n)
return output
There isn’t a lot to say about this implementation. No pre-allocation, iterate over the x’s and y’s which represent points in the complex plane to create a complex number c then compute the series until we reach the max iterations or it diverges. If we time it we get 19.78 seconds total runtime. By profiling it line by line we see the following:
Line # Hits Time Per Hit % Time Line Contents
==============================================================
12 @profile
13 def mandelbrot_naive(xs, ys, _, max_iter, power=2):
14 1 1.0 1.0 0.0 output = []
15
16 1201 275.0 0.2 0.0 for i in range(len(xs)):
17 1441200 247858.0 0.2 0.3 for j in range(len(ys)):
18 1440000 335255.0 0.2 0.4 c = complex(xs[i], ys[j])
19 1440000 176060.0 0.1 0.2 n = 0
20 1440000 183386.0 0.1 0.2 z = 0
21 157387973 34612134.0 0.2 41.8 while abs(z) < 2 and n < max_iter:
22 155947973 26493423.0 0.2 32.0 z = z**power + c
23 155947973 20620193.0 0.1 24.9 n += 1
24 1440000 177280.0 0.1 0.2 output.append(n)
25 1 1.0 1.0 0.0 return output
There are a few minor improvements we can make here, first we can pre-allocate the list by using a list comprehension which lets us do one loop that is len(xs) * len(ys) big. Doing so reduces the time to 19.62 seconds, which since its not a drastic improvement I will not show the line profile. Furthermore, timing the two methods many times there is not a clear improvement here. This simple approach can be rendered with Pillow by using an HSV transformation of the iterations values. We want 1000 iterations to be black and everything else to be not-black with a reasonable scheme. How to visualize this has a lot of different algorithms, but with this implementation I just did a simple scaling. By doing so we get this image:
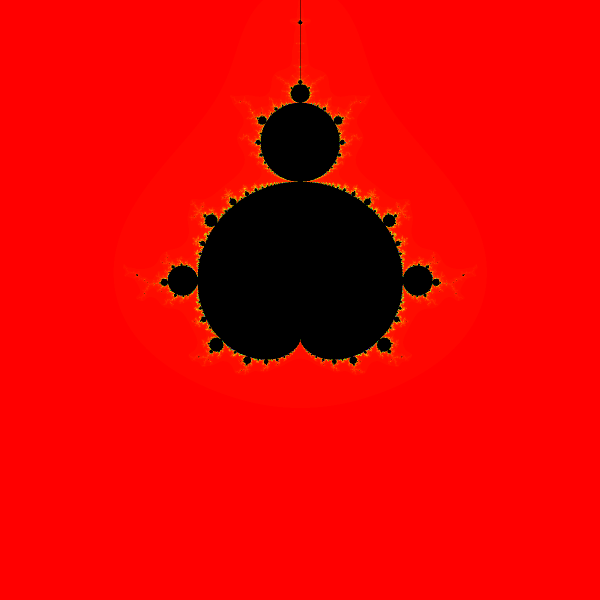
With Numpy Link to heading
Since we are computing all values across the range of the complex plane xs by ys, we can try just doing a simple conversion to NumPy. This allows us to compute all of the numbers at once. Below is that implementation:
def mandelbrot_numpy(xs, ys, c, max_iter, power=2):
# calculate z using numpy, this is the original
# routine from vegaseat's URL
x, y = np.array(xs), np.array(ys)
c = np.ravel(x + y[:, None] * 1j)
output = np.resize(
np.array(
0,
),
c.shape,
)
z = np.zeros(c.shape, np.complex64)
for it in range(max_iter):
z = np.power(z, power) + c
done = np.greater((z.real * z.real) + (z.imag * z.imag), 4.0)
c = np.where(done, 0 + 0j, c)
z = np.where(done, 0 + 0j, z)
output = np.where(done, it, output)
return output
Note that we have to do a bit of book-keeping each run to mark entries in the output as done, if they have exceeded the threshold for divergence. This method drops the runtime to 11.62s given the same parameters. This is a 41% reduction from our original implementation.
Looking at the profile we see:
Line # Hits Time Per Hit % Time Line Contents
==============================================================
84 @profile
85 def mandelbrot_numpy(xs, ys, c, max_iter, power=2):
86 # calculate z using numpy, this is the original
87 # routine from vegaseat's URL
88 1 65.0 65.0 0.0 x, y = np.array(xs), np.array(ys)
89 1 9814.0 9814.0 0.1 c = np.ravel(x + y[:, None] * 1j)
90 2 261980.0 130990.0 3.0 output = np.resize(
91 2 2.0 1.0 0.0 np.array(
92 1 0.0 0.0 0.0 0,
93 ),
94 1 0.0 0.0 0.0 c.shape,
95 )
96 1 76.0 76.0 0.0 z = np.zeros(c.shape, np.complex64)
97 1001 602.0 0.6 0.0 for it in range(max_iter):
98 1000 4427647.0 4427.6 50.1 z = np.power(z, power) + c
99 1000 1819997.0 1820.0 20.6 done = np.greater((z.real * z.real) + (z.imag * z.imag), 4.0)
100 1000 826375.0 826.4 9.4 c = np.where(done, 0 + 0j, c)
101 1000 785184.0 785.2 8.9 z = np.where(done, 0 + 0j, z)
102 1000 701267.0 701.3 7.9 output = np.where(done, it, output)
103 1 1.0 1.0 0.0 return output
This is essentially what we saw in the naive implementation, all the time is spent computing the next z and checking if its done. There may be other optimizations we could do here with just straight Python + NumPy, but it would be a lot of work with an uncertain outcome. Instead let’s look at some compiled options for doing the same thing.
Numba Magic Link to heading
Numba is a Python + NumPy JIT compiler. It does not work with all of Python or NumPy and their are a lot of common libraries it does not work with, like SciPy. However, when your code is doing lots of loops or NumPy manipulations you can get a lot of performance improvements with very little effort. If we take our second implementation and add the Numba @jit annotation to it, Numba will compile it giving us a huge speed boost. That looks like this:
@jit(nopython=True, cache=True)
def mandelbrot_numba(xs, ys, _, max_iter, power=2):
cs = [complex(x, y) for x in xs for y in ys]
output = [0 for _ in range(len(cs))]
for i, c in enumerate(cs):
n = 0
z = 0
while abs(z) < 2 and n < max_iter:
z = z**power + c
n += 1
output[i] = n
return output
The @jit annotation tells Numba to compile this function just in time. The nopython=True says that for this function we do not want to allow calling back into the Python interpreter. If you have code that this will not work with Numba will fail when you go to run the function. If at all possible we want nopython to be set to True, as not doing so is a big performance hit. The @jit annotation and nopython=True parameter can be shortened to @njit. The cache=True parameter says that after this function is compiled the first time we would like to cache that version, this has a few gotchas but will prevent us from having to compile each new program instance. By default the first time a Numba annotated function is called it will be compiled, thereafter using the compiled version. This means that the first call should be slower.
Using Numba drops our runtime to 1.5s! This is a 92% reduction in speed by just using an annotation with Numba. Again this approach will not work for everything, but where it does work the gains are significant for very little effort. One warning is that Numba does not use NumPy directly instead it replicates the NumPy api. In practice the implementations produce the same results, but validate your results to be sure.
Cython, a little compiled goodness Link to heading
At this point the results with Numba are so good that in the real world I would likely stop there. For learning though I decided to reimplement this using Cython. Cython is a static compiler for Python and the extended Cython language. Cython is a superset of Python adding type annotations, support for calling C functions, and other tricks. Cython makes it easy to call C code, as well as turn your existing Python into compiled C with very little effort. Here is my Cython implementation of computing the Mandelbrot set:
from cython.parallel import prange
import numpy as np
cimport numpy as np
def calcualte_z(double complex[:] cs, int max_iter, int power):
# Cython version of calcualting the mandelbrot set
cdef unsigned int i, length
cdef double complex z, c
cdef int[:] output = np.empty(len(cs), dtype=np.int32)
length = len(cs)
#using nogil as we are using prange from openmp and this area does not call into the python interpretter until we are done
with nogil:
#this uses openmp to paraellelize out across all of the complex numbers we have, schedule='guided' handles breaking the tasks up nicely
for i in prange(length, schedule='guided'):
z = 0
output[i] = 0
c = cs[i]
while output[i] < max_iter and (z.real * z.real + z.imag * z.imag) < 4:
z = z * z + c
output[i] += 1
return output
As you can see most of this looks like the existing Python implementation. I have added in types for all parameters and variables. The double complex[:] is a Cython typed memory view, which is one way of defining a NumPy array variable. If you have multi-dimension arrays you would add a ,: for each multi-dimension. The cdef keyword says that we are defining a c type, similarly cimport specifically imports the C NumPy api. We use nogil to indicate that this block of code will not call into the Python interpreter, which removes the gil threading restrictions. In practice this is done for either parallelization or occasionally long running io code. In this case we are using it to leverage OpenMP to parallelize the computation of each complex number we are considering. Cython exposes OpenMP through the prange function. Here we are trusting OpenMP to smartly schedule threads with chunks of work. Past this the code is about the same as the other implementations. To call this from Python we do the following:
import cfractals
def mandelbrot_cython(xs, ys, _, max_iter, power=2):
cs = np.array([complex(x, y) for x in xs for y in ys])
output = cfractals.calcualte_z(cs, max_iter, power)
return output
To actually use this code we have to first compile it. In this case I decided to use setuptools, here is my setup.py:
from setuptools import setup
from setuptools import Extension
import numpy as np
ext_modules = [Extension("cfractals", ["cfractals.pyx"], extra_compile_args=['-fopenmp', '-I/opt/homebrew/Cellar/libomp/18.1.0/include/omp.h', '-I/opt/homebrew/Cellar/libomp/18.1.0/lib'], extra_link_args=['-fopenmp'])]
from Cython.Build import cythonize
setup(ext_modules=cythonize(ext_modules, compiler_directives={"language_level": 3}), include_dirs=np.get_include())
Since I am using OpenMP I had to add the correct includes, for the steps I took to get this on my machine see the repo above’s README. Additionally, I would not expect this to build on another OS, without changing the compile flags. For our troubles of going through this at 1200x1200 images with 1000 iterations the results are comparable to Numba. However at larger sizes / iterations the Cython implementation wins.
Redoing everything in C Link to heading
I wanted to see how close to C performance all of the optimizations I did ended up being. I haven’t written any C since I was in college, but no time like the present. I just went with a naive implementation, using a flattened 2d array to keep things simple.
double *mandelbrot(long double *xs, long double *ys, int height, int width, int maxIter) {
double *iterations = malloc(height * width * sizeof(double));
// this is invoking OpenMP so that we get parallelizm, smartly
#pragma omp parallel shared(iterations) shared(xs) shared(ys) shared(height) shared(width) shared(maxIter)
{
#pragma omp for collapse(2)
for (int y = 0; y < height; y++) {
for (int x = 0; x < width; x++) {
long double complex c = xs[x] + I * ys[y];
int n = 0;
long double complex z = 0;
while (cabsl(z) < 2 && n < maxIter) {
z = z * z + c;
n += 1;
}
if (n == maxIter) {
iterations[y * width + x] = (double)n;
} else {
iterations[y * width + x] = n + 1 - clog(clog(cabsl(z)));
}
}
}
}
return iterations;
}
The only thing here that may not be obvious is the OMP collapse clause. This is used to smash the nested forloop into a single loop and parallelize each item in it. My C isn’t great so I am probably being bad with my lack of a free call in the calling code. The calling code is using raylib’s rpng library to render the Mandelbrot set after we find the values. If we time this implementation on average it works out to about 3 seconds. However, if we remove the png output so that we are only timing the Mandelbrot set generation then we end up with an average of 0.9 seconds. I didn’t feel like finding a png library that would be able to compete with my Python implementation, but I am happy that the C version is faster than the Python implementation (if I exclude both png renderings).
I did spend a bit more time on the rendering algorithm here, which resulted in a prettier Fractal image. As can be seen below:
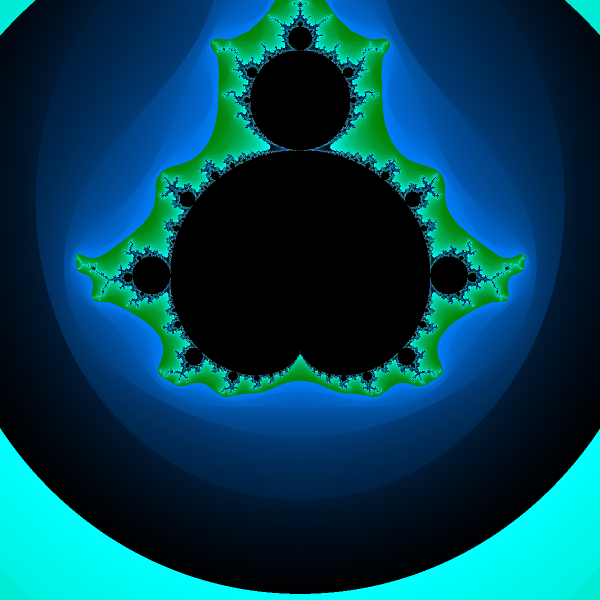
Fractal zooming Link to heading
I was still enjoying playing with fractals and C. So I decided to implement a Mandelbrot set visualizer using raylib. The core of this code still uses the mandelbrot function from above, but in addition I am automatically zooming in on an area that is on the edge of the Mandelbrot set. Below is the code I use to render the Mandelbrot set after recalculating it at each step.
void renderMandelbrot(double *fractals, int height, int width, struct ZoomSeed *zs) {
int stepsSinceM = 10000;
for (int y = 0; y < height; y++) {
for (int x = 0; x < width; x++) {
double iterations = fractals[y * width + x];
Color color;
stepsSinceM += 1;
if ((int)iterations == MAXITER) {
color = BLACK;
stepsSinceM = 0;
} else {
if (zs->iterations < iterations && stepsSinceM < 10) {
zs->iterations = iterations;
zs->x = x;
zs->y = y;
zs->rounds = 0;
}
int r = (int)floor((float)iterations / (float)MAXITER * 255);
int g = (int)floor((log(iterations) + 1.0 - (float)iterations / (float)MAXITER) / 2.0 * 255);
int b = (int)floorl((log(iterations) + (1.0 - (float)iterations / (float)MAXITER)) * 255);
color = (Color){r, g, b, 255};
}
DrawPixel(x, y, color);
}
DrawPixel(0, 0, RED);
}
zs->rounds += 1;
}
The ZoomSeed struct that is passed in is then used to adjust the Mandelbrot set that is calculated in the next step, centering the field of view on x/y with a window size 0.95 of the previous window, that way we are always zooming into a new area. This simple approach does cause the view to jump around a bit, but overall it leads to a really cool way of visualizing the Mandelbrot set.
Conclusion Link to heading
This is a simple problem to implement, but it shows how drastic speed improvements can be made in Python by leveraging libraries that produce compiled code. Not all code should be optimized, but if its a noticibly slow area of your code consider Numpy, Numba, and Cython. Or just throw away Python and rewrite in C.